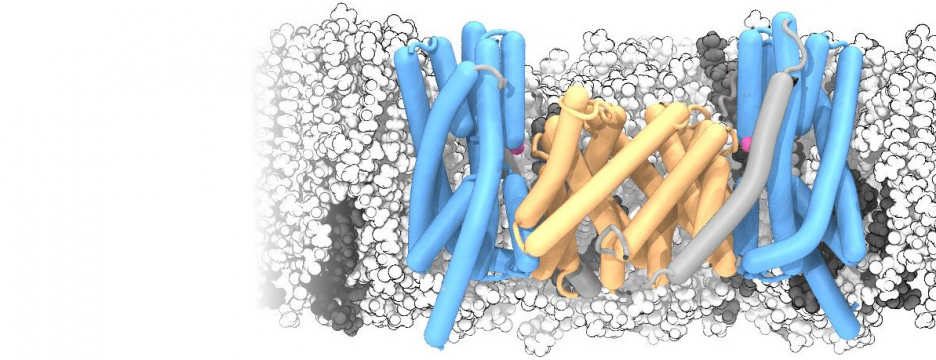
Simulations of membrane proteins
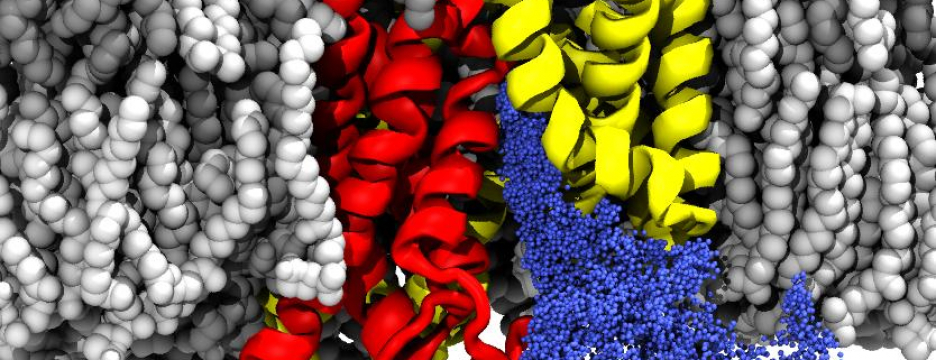
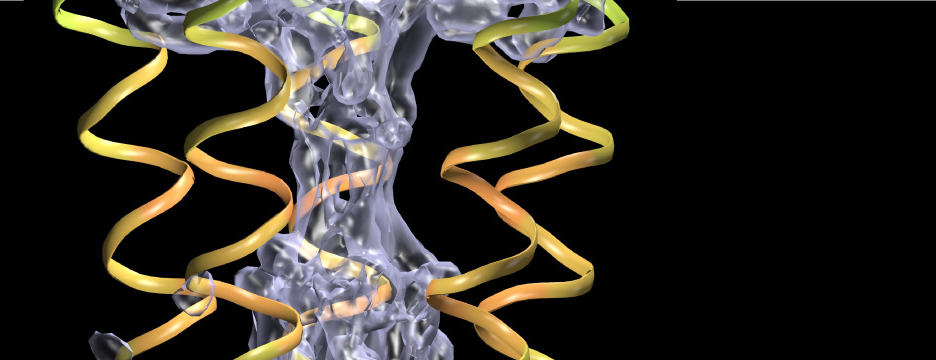
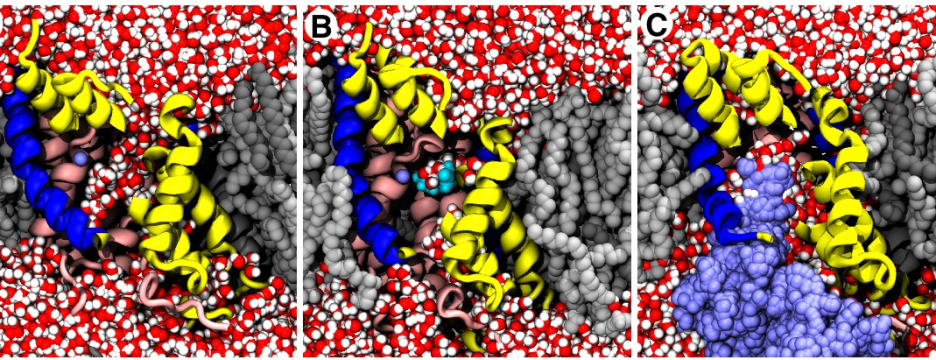
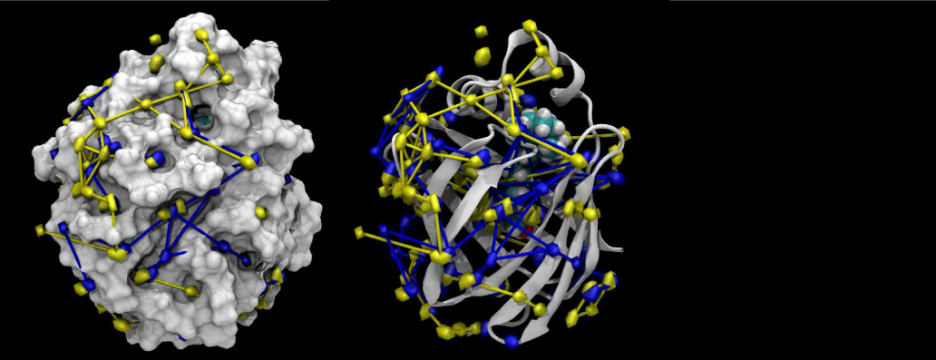
Many proteins in the living cell can be understood as molecular machines that use a source of energy to produce mechanical or chemical work. My lab’s primary interest is in those proteins located in the cell membrane that move nutrients, signalling molecules, or waste products into and out of the cell. We study their molecular mechanisms of action by detailed molecular dynamics simulations, which provide a “movie” of full atomic detail of a working protein.
Current areas of interest focus on the mechanisms of secondary active transport; methods to accurately simulate macromolecular transitions that are crucial in understanding ligand binding, gating in ion channels, or the translocation of substrates through the cell membrane; and the role of water in confined geometries, for instance in ion channel gating mechanisms, ligand discrimination, or drug binding.