X-ray crystallography and Simulations
Structures of membrane proteins can be obtained by the experimental technique of X-ray crystallography. However, proteins are typically not crystallized in their native environment, the lipid membrane. Molecular dynamics simulations of the protein in the membrane provide a realistic model of the interactions between transporter and lipid bilayer.
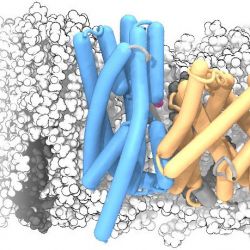
The structure of a membrane protein, the sodium/proton antiporter NapA, was solved by X-ray crystallography by our collaborators. In order to better understand its interaction with the membrane we simulated the NapA dimer in a POPE:POPC membrane. The dimerization domain is shown in gold while the core domains are depicted in electric blue. During the simulation, two sodium ions (magenta) spontaneously bound to the putative ion binding sites near the center of each protomer.
The helices were drawn with the Bendix plugin for VMD and rendered with Tachyon raytracer.
The story behind this image is described under A two-domain elevator mechanism for sodium/proton antiport.

