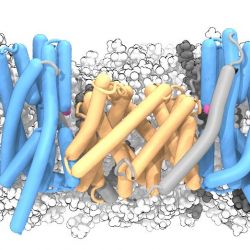
A two-domain elevator mechanism for sodium/proton antiport
Organisms from bacteria to man must control the pH and salt content of cells to function properly. They achieve this by actively pumping either protons or sodium ions out of the cell, using a special class of proteins that sit in the cell membrane, the so called “sodium-proton exchangers”. Dysfunction of these exchanger proteins in humans has been linked to diseases such as hypertension, heart failure, epilepsy and autism. We are interested in the molecular mechanism of these transporter proteins in order to better understand their failure to function in diseases (e.g. due to mutations) and to ultimately develop drugs that specifically interact with the transporter.
In a paper published in Nature1 we describe the conformation of the sodium/proton antiporter NapA in an outward facing conformation (PDB ID 4BWZ) from an X-ray crystal structure at 3 Å resolution. Together with the inward facing conformation known from another member of the
sodium/proton antiporter family, the homologous protein NhaA2 (PDB ID 4AU5 and 4ATV and also 1ZCD ; see our work on the crystal structure of the sodium-proton antiporter NhaA dimer), we can now for the first time see the conformational change that the protein has to undergo when transporting protons into the cell and sodium ions out of the cell.
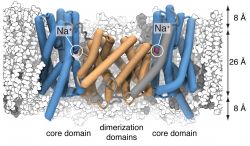
The sodium/proton antiporter NapA was simulated in a POPE (white):POPC (gray) membrane. The dimerization domain is shown in orange while the core domains are depicted in blue. During the simulation, two sodium ions (magenta) spontaneously bound to the putative ion binding sites near the center of each protomer.
Sodium/proton antiporters are fast for active transport proteins; depending on conditions they can go through up to 1500 transport cycles per second. It was thought that such fast transport would be reflected in relatively small and localized conformational changes. However, the structures show that the actual conformational change is rather large, whereby the so-called core domain (blue in the figure) moves relative to the dimer domain (orange). In this way, conserved aspartate residues are exposed to either the extracellular (periplasmic) or intracellular (cytosolic) side of the membrane, in accordance with the alternating access mechanism of transmembrane transport.
Our molecular dynamics simulations identified the conserved Asp157 as the most likely binding site for a sodium ion. The simulations showed multiple binding and unbinding events for the deprotonated (charged) sidechain. However, protonating Asp157 abolished ion binding, an observation in line with recent experimental measurements that conclude that Na+ and H+ compete for the same binding site3.
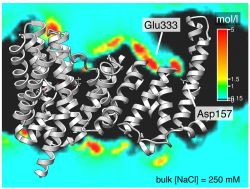
The sodium/proton antiporter NapA was simulated in a 250 mM NaCl solution. The spatial density of sodium ions was sliced through and overlaid on the structure. Very high Na concentrations (greater than 5 M) are visible near the conserved Asp157 (negatively charged) and Glu333 (which takes the functional position of Asp133 in the homologous protein NhaA).
Analysing the density of Na+ ions over the course of the simulations (see figure above) clearly shows that sodium ions are concentrated near Asp157 and also near Glu333. The latter is interesting because the charged carboxyl group occupies almost the same position in space as the carboxyl group of Asp133 in NhaA.
In summary, we have made an important step towards unraveling the molecular mechanism of sodium/proton antiport even though a number of crucial questions remain open: What is the nature of the conformational change, how is it triggered, which residues participate in proton binding, and how is the mechanism affected by mutations or binding of small molecules.
References
1 C. Lee, H.J. Kang, C. von Ballmoos, S. Newstead, P. Uzdavinys, D.L. Dotson, S. Iwata, O. Beckstein, A.D. Cameron, and D. Drew. A two-domain elevator mechanism for sodium/proton antiport. Nature 501 (2013), 573–577. doi: 10.1038/nature12484
2 C. Lee, S. Yashiro, D.L. Dotson, P. Uzdavinys, S. Iwata, M.S.P. Sansom, C. von Ballmoos, O. Beckstein, D. Drew, and A.D. Cameron. Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights. J Gen Physiol 144 (2014), 529-544. doi: 10.1085/jgp.201411219 (ASU repository item 27919 )
3 T. Mager, A. Rimon, E. Padan, and K. Fendler. Transport mechanism and pH regulation of the Na+/H+ antiporter NhaA from Escherichia coli: an electrophysiological study. J Biol Chem, 286 (2011), 23570–23581. doi: 10.1074/jbc.M111.230235


Discuss: “A two-domain elevator mechanism for sodium/proton antiport”