
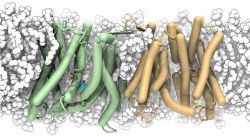
Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights
Sodium–proton antiporters rapidly exchange protons and sodium ions across the membrane to regulate intracellular pH, cell volume, and sodium concentration. Their failure in humans is implicated in epilepsy and hypertension, and they are therefore common drug targets. Despite decades of study, however, how ion binding and release is coupled to the conformational changes associated with transport is still not clear.
In a paper published in the Journal of General Physiology1 (and featured on the cover), we report a crystal form of the prototypical sodium-proton antiporter NhaA from Escherichia coli in which the protein is seen as a dimer (PDB ID 4AU5 and 4ATV). This new structure features a salt bridge between an essential aspartic acid (Asp163) and a conserved lysine (Lys300). An equivalent salt bridge is present in the homologous transporter NapA2 (PDB ID 4BWZ), but not in the only other known crystal structure of NhaA3 (PDB ID 1ZCD), which provides the foundation of most existing structural models of electrogenic sodium–proton antiport.
We performed molecular dynamics simulations of the protein dimer in a fully-atomistic membrane with explicit water and ions. These simulations showed that the stability of the salt bridge is weakened by sodium ions binding to Asp164 and the neighboring Asp163. This suggests that the transport mechanism involves Asp163 switching between forming a salt bridge with Lys300 and interacting with the sodium ion.
We also performed simulations with differing protionation states for the conserved residues. In cases where Asp164 is protonated, no sodium binding is observed. When Asp163 is protonated or Lys300 is deprotonated, the stability of the salt bridge is weakened. pKa calculations suggest that Asp163 is highly unlikely to be protonated when involved in the salt bridge, which suggests a possible role for Lys300 in proton binding.
As it has been previously suggested that Asp163 is one of the two residues through which proton transport occurs, these results have clear implications to the current mechanistic models of sodium–proton antiport in NhaA.
References
1 C. Lee, S. Yashiro, D.L. Dotson, P. Uzdavinys, S. Iwata, M.S.P. Sansom, C. von Ballmoos, O. Beckstein, D. Drew, and A.D. Cameron. Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights. J Gen Physiol 144 (2014), 529-544. doi: 10.1085/jgp.201411219 (ASU repository item 27919 )
2 C. Lee, H.J. Kang, C. von Ballmoos, S. Newstead, P. Uzdavinys, D.L. Dotson, S. Iwata, O. Beckstein, A.D. Cameron, and D. Drew. A two-domain elevator mechanism for sodium/proton antiport. Nature 501 (2013), 573–577. doi: 10.1038/nature12484
3 C. Hunte, E. Screpanti, M. Venturi, A. Rimon, E. Padan, and H. Michel. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435 (2005), 1197–1202. doi: 10.1038/nature03692


Discuss: “Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights”