Molecular mechanism of ligand recognition by the Mhp1 transporter
Mhp1 is a nucleobase-sodium symporter and member of the NCS1 family of transporters 1. It couples the energetically favorable uptake of sodium to the uptake of hydantoin derivatives against their chemical transmembrane gradient. Structurally, it shares a five helix inverted repeat architecture with a large number of other transporters such as LeuT and vSGLT2. We previously showed through a combination of X-ray crystallography and computer simulations that Mhp1 functions according to the alternating access mechanism 3. However, the resolution of the previous structures had not been sufficient to understand how substrate molecules (5-substituted hydantoins) bind to the transporter and what interactions are responsible for the selectivity of the transporter. Through a combination of crystallography, molecular dynamics, site?directed mutagenesis, biochemical/biophysical assays, and the design and synthesis of novel ligands we were recently able to define and measure the determinants of substrate selectivity and propose a refined model of the transport process 4.
The following summarizes some of the important findings from the article while highlighting connections to our previous work and how MD simulations contributed to unraveling The molecular mechanism of ligand recognition by membrane transport protein, Mhp1 4.
Conformation of the ligand
The only ligand-bound structure of Mhp1 was the 4-Å resolution outward-facing occluded structure (PDB 2JLO) with the substrate L-benzylhydantoin (abbreviated as L-BH) bound. Based on the small-molecule crystal structure 5 (CCSD deposition number 659125, CCSD code CIJYOK), L-BH was modeled into the density in a “U-shaped” or “folded-over” (g+) conformation, which is also consistent with solution NMR studies that showed the folded-over conformation to be the pre-dominant one 6. In our new crystal structure with L-indolyl-methyl-5-hydantoin (L-IMH) bound (PDB ID 4D1A) , however, the density of the ligand suggested a more extended conformation (g–). MD simulations showed that the L-BH and L-IMH molecules in solutions can interconvert between folded-over and extended conformations approximately on the 10-ns time scale. For L-BH, the folded-over conformation is indeed more stable as indicated by the earlier NMR experiments. However, bound to Mhp1, folded-over L-BH (g+) adopts the extended conformation (g–) within less than 50 ns in repeated simulations. We were able to obtain a improved crystal structure with L-BH bound (at 3.8 Å resolution, 4D1B), which now shows L-BH in the extended conformation7, in agreement with the MD. L-IMH remains extended in MD simulations.
Determinants of selectivity
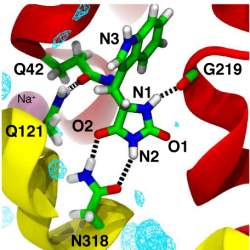
Mhp1 discriminates ligands by requiring two hydrogen bonds to the polar hydantoin moiety: one is provided by the sidechain of the conserved Asn318 and the other by the hitherto unrecognized backbone carbonyl of Gly219. This became particular clear from MD simulations of two transported substrates, L-IMH and L-BH. Multiple MD simulations of length 100 ns or longer with different starting conformations of the ligands were performed. The hydrogen bonds between chemically equivalent atoms (including solvent atoms) were analyzed with MDAnalysis and clustered. The clustered heatmap (subfigure c below) reveals the hydrogend bonding pattern and provides a fingerprint of hydantoin-protein interactions. It clearly identifies the hydrogen bonds to G219 and N318 as the most commonly observed ones across simulations.
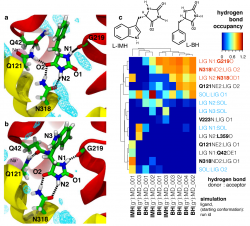
Analysis of hydrogen bonds between Mhp1 and two substrate molecules via MD simulations. The ligand and important residues are shown as sticks with hydrogen bonds as broken black lines. Helices TM1 and TM6 from the bundle are in red and TM3 and TM8 from the hash motif in yellow (parts of TM3 were removed for clarity); a sodium ion in the Na2 site is visible in the background. Water density is shown as a cyan mesh, contoured at 1.5 times the bulk value. Equivalent atoms on the ligands are labelled. (a) L-benzylhydantoin (L?BH). (b) L-indolylmethyl-hydantoin (L?IMH). (c) Clustered fingerprint analysis of hydrogen bonds. The occupancy (average number of hydrogen bonds between ligand atoms and protein or solvent atoms) from all MD simulations was clustered to show the most commonly occurring hydrogen bonding patterns.
However, transport and binding assays showed that these hydrogen bonds are not sufficient to determine selectivity because hydantoin itself and other hydantoin derivatives such as allantoin (which has a polar group in the 5 position of the hydantoin ring) neither bind nor are transported. Instead, a bulky hydrophobic substituent such as a phenyl group (resulting in L-benzylhydantoin) or an indole moiety (as for the best transported known substrate L-indolyl-methyl-5-hydantoin ) is also required. Crystal structures confirmed docking calculations that show that the hydrophobic group fits into a sizable hydrophobic pocket.
But not just any hydrophobic group will do: The hydrophobic moiety has to be the right size: if the substituent is too large such as a naphtyl group then the molecule binds very strongly but is not transported. In fact, 5-(2-naphtylmethyl)-L-hydantoin binds stronger than any of the other transported substrates and acts as a competitive inhibitor.
Coupling of substrate binding to conformational changes
A comparison of the crystal structure of L-NMH bound to Mhp1 (PDB 4D1D) to the apo structure (2JLN) and the L-IMH bound one (4D1A) as show in the figure, revealed the reason why L-NMH is not transported.
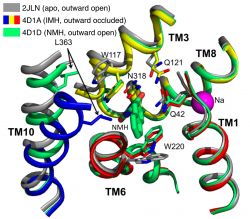
Comparison of structures of Mhp1 in the apo state and with L-indolylmethyl-5-hydantoin (IMH) and L-naphylmethyl-5-hydantoin (NMH) bound. The NMH-bound structure contains the bound ligand but helix TM10 remains in the open position (similar to the outward-facing open ligand-free (apo) structure), due to steric hindrance between the bulky sidechain of Leu363 and the naphtyl moiety of the NMH ligand.
As we showed previously 3, Mhp1 transitions on substrate binding from an outward-facing open conformation to an occluded conformation by a movement of the N-terminal half of helix TM10 (the so-called extracellular gate). However, the L-NMH bound structure shows TM10 in the open conformation, i.e. the gate remains open. The reason is that the bulky sidechain of Leu363 clashes with the large naphtyl moiety and the ligand thus wedges the transporter open and stabilizes it in the outward facing open conformation. When the steric clash was removed by mutating L363 to Ala (L363A) then L-NMH was transported at least as well as L-IMH, thus confirming that the size of the hydrophobic pocket contributes to the selectivity. Interestingly, this also strongly supports our previous hypothesis that extracellular gate closure is mechanically coupled to the large conformational change of switching from the outward facing conformation to the inward facing one 3 because transport was inhibited while the extracellular gate remained open.
References
- a S. Weyand, P. Ma, M. Saidijam, J. Baldwin, O. Beckstein, S. Jackson, S. Suzuki, S. G. Patching, T. Shimamura, M. S. P. Sansom, S. Iwata, A. D. Cameron, S. A. Baldwin, and P. J. F. Henderson. The nucleobase-cation-symport-1 family of membrane transport proteins. In A. Messerschmidt, editor, Handbook of Metalloproteins. Wiley, 2010. doi: 10.1002/0470028637.met268
- a A. D. Cameron, O. Beckstein, and P. J. Henderson. The five-helix inverted repeat superfamily of membrane transport proteins. In G. Roberts, editor, Encylopedia of Biophysics. Springer, 2013. pp 1481—1485. doi: 10.1007/978-3-642-16712-6_772
- a b c T. Shimamura, S. Weyand, O. Beckstein, N. G. Rutherford, J. M. Hadden, D. Sharples, M. S. P. Sansom, S. Iwata, P. J. F. Henderson, and A. D. Cameron. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science, 328 (5977):470–473, 2010. doi: 10.1126/ science.1186303 .
- a b K. J. Simmons, S. M. Jackson, F. Brückner, S. G. Patching, O. Beckstein, E. Ivanova, T. Geng, S. Weyand, D. Drew, J. Lannigan, D. J. Sharples, M. S. P. Sansom, S. Iwata, C. W. Fishwick, A. P. Johnson, A. D. Cameron, and P. J. Henderson. The molecular mechanism of ligand recognition by membrane transport protein, Mhp1. EMBO J, 33 :1831–1844, 2014. doi: 10.15252/embj.201387557 .
- a G. E. Delgado, A. J. Mora, J. Uzcátegui, A. Bahsas, and A. Briceño. (S)-5-Benzylimidazolidine-2,4-dione monohydrate. Acta Crystallographica Section C-Crystal Structure Communications, 63 :O448–O450, 2007. doi: 10.1107/S0108270107028090
- a E. Kleinpeter. The structure of hydantoins in solution and in the solid state. Structural Chemistry, 8 (2): 161–173, 1997. doi: 10.1007/BF02262852.
- a The new L-BH-bound structure 4D1B (3.8 Å resolution, extended L-BH conformer, g–) now supersedes the old structure 2JLO (4.0 Å resolution, folded-over g+ L-BH conformer).


Discuss: “Molecular mechanism of ligand recognition by the Mhp1 transporter”