AdK apo PMF
Adenylate kinase (AdK) is a enzyme that undergoes a large hinge-like motion. Because of an abundance of structural and functional data, it has become a standard system to test computational methods for sampling conformational transitions.1
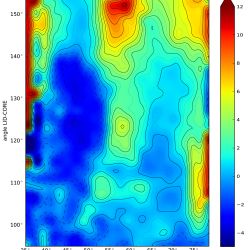
In 2009, we studied the conformational transition between open and closed E. coli AdK without substrate, i.e. “apo AdK”, with a variety of computational methods.2 As part of the study we also produced a free energy landscape (a potential of mean force or PMF) as a function of two collective variables, the angles formed by the LID and NMP domains with the CORE domain.3 We 2 and others4,5 have used this PMF to compare methods that sample transition paths to the underlying free energy landscape.
We make the raw data of the underlying free energy landscape publicly available to other researchers so that they can use them in their own research.
Terms of Use
The data are made available under a Attribution-ShareAlike 4.0 International licence (include the following when using the data):
 Adenylate Kinase Potential of Mean Force by O Beckstein, EJ Denning, JR Perilla, TB Woolf is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. Based on a work at http://becksteinlab.physics.asu.edu/file_download/11/free_bw2_tol1e-05.dat.
Adenylate Kinase Potential of Mean Force by O Beckstein, EJ Denning, JR Perilla, TB Woolf is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. Based on a work at http://becksteinlab.physics.asu.edu/file_download/11/free_bw2_tol1e-05.dat.
When you make use of the data (contained in the file free_bw2_tol1e-05.dat) in published work, cite the paper2
O. Beckstein, E. J. Denning, J. R. Perilla, and T. B. Woolf. Zipping and unzipping of adenylate kinase: Atomistic insights into the ensemble of open ? closed transitions. J. Mol. Biol., 394(1):160–176, 2009.
Data 
The file free_bw2_tol1e-05.dat contains the PMF data shown in Fig. 4a of the JMB paper2.
The image shows the data plotted with cubic spline smoothing.
File format
free_bw2_tol1e-05.dat is the output from WHAM. The important data columns are
- NMP-core angle (degrees)
- LID-core angle (degrees)
- free energy in kcal/mol
(Other columns are output from wham and can be ignored.)
Methods
Conformations of E. coli AdK were umbrella-sampled in the space of the NMP-core and LID-core angles.3 The protein was modelled in implicit solvent with the ACE2 electrostatics model. The resulting umbrella data were unbiased using Alan Grossfield’s wham code with
- bin size 2º
- tolerance of the self consistency procedure 1e-5 kT
- limits 34º < NMP < 80º and 94º < LID < 156º
The first 2000 frames (200ps) of each window were discarded as equilibration and the remaining 3000 frames were used for the PMF. For further details please see the paper.2
References
- a S. L. Seyler and O. Beckstein, O. Sampling large conformational transitions: adenylate kinase as a testing ground. Mol. Simul., 40(10–11): 855–877, 2014.
- a b c d e f O. Beckstein, E. J. Denning, J. R. Perilla, and T. B. Woolf. Zipping and unzipping of adenylate kinase: Atomistic insights into the ensemble of open / closed transitions. J. Mol. Biol., 394(1):160–176, 2009.
- a b See the 2009 paper2 for the definitions and the MDAnalysis tutorial’s Exercise 4 for Python code to calculate the angles.
- a M. Gur, J. D. Madura, and I. Bahar. Global transitions of proteins explored by a multiscale hybrid methodology: Application to adenylate kinase Biophysical Journal, 105(7):1643 – 1652, 2013.
- a A. Uyar, N. Kantarci-Carsibasi, T. Haliloglu, and P. Doruker. Features of large hinge-bending conformational transitions. Prediction of closed structure from open state. Biophysical Journal, 106(12):2656 – 2666, 2014



Discuss: “AdK apo PMF”