Atomically detailed molecular dynamics simulations
Using molecular dynamics (MD) computer simulations we can study membrane proteins in atomic detail, down to the movements of individual water molecules (red/white in the image), ions (blue), lipids (gray), and the protein itself.
Alternating access in Mhp1 describes in more detail what one can learn from MD simulations of transporters.
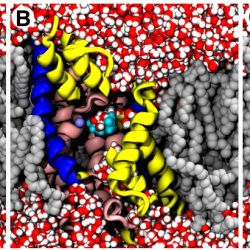
The image shows the accessibility of the binding sites of Mhp1 in three conformational states. Computer simulations of the Mhp1 transport protein (helices in cartoon representation, front cut away) in a lipid membrane (gray) were carried out to determine the functional states of three crystallographic structures of Mhp1 (A: outward facing open, 2JLN; B: outward facing occluded, 2JLO, and C: inward facing open 2X79). Water molecules (red/white) can only access the substrate and sodium binding site in the open conformations, and the binding sites are sealed in the occluded state. This is the behaviour is predicted by the alternating access mechanism. Repeated simulations showed that inward-facing conformation (C) readily released the bound Na+ ion; the positions of the ion (magenta spheres) from six simulations were all overlaid in panel C and define the exit pathway.


