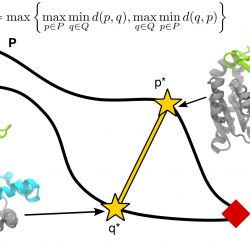
Sampling macromolecular transitions
A fundamental problem in computational biophysics is to deduce the function of a protein from the structure. Many biological macromolecules such as enzymes, molecular motors, or membrane transport proteins perform their function by cycling between multiple conformational states. Understanding such conformational transitions, which typically occur on the millisecond to second time scale, is central to understanding protein function. Molecular dynamics (MD) computer simulations have become an important tool to connect molecular structure to function but equilibrium MD simulations are rarely able to sample on time scales longer than a few microseconds—orders of magnitudes shorter than the time scales of interest.
A range of different simulation methods have been proposed to overcome the time-scale limitation of simulating conformational changes. These include calculations of the free energy landscape and path sampling methods to directly sample transitions between known conformations. All these methods solve the problem to sample infrequently occupied but important regions of configuration space. Many path-sampling algorithms have been applied to the closed <—> open transition of the enzyme adenylate kinase (AdK), which undergoes a large, clamshell-like conformational transition between an open and a closed state. In our review Sampling large conformational transitions: adenylate kinase as a testing ground1 we examine approaches to sampling macromolecular transitions through the lens of AdK, focusing our main discussion on the current state of knowledge — both from simulations and experiments — about the transition pathways of ligand-free AdK, its energy landscape, transition rates, and interactions with substrates.
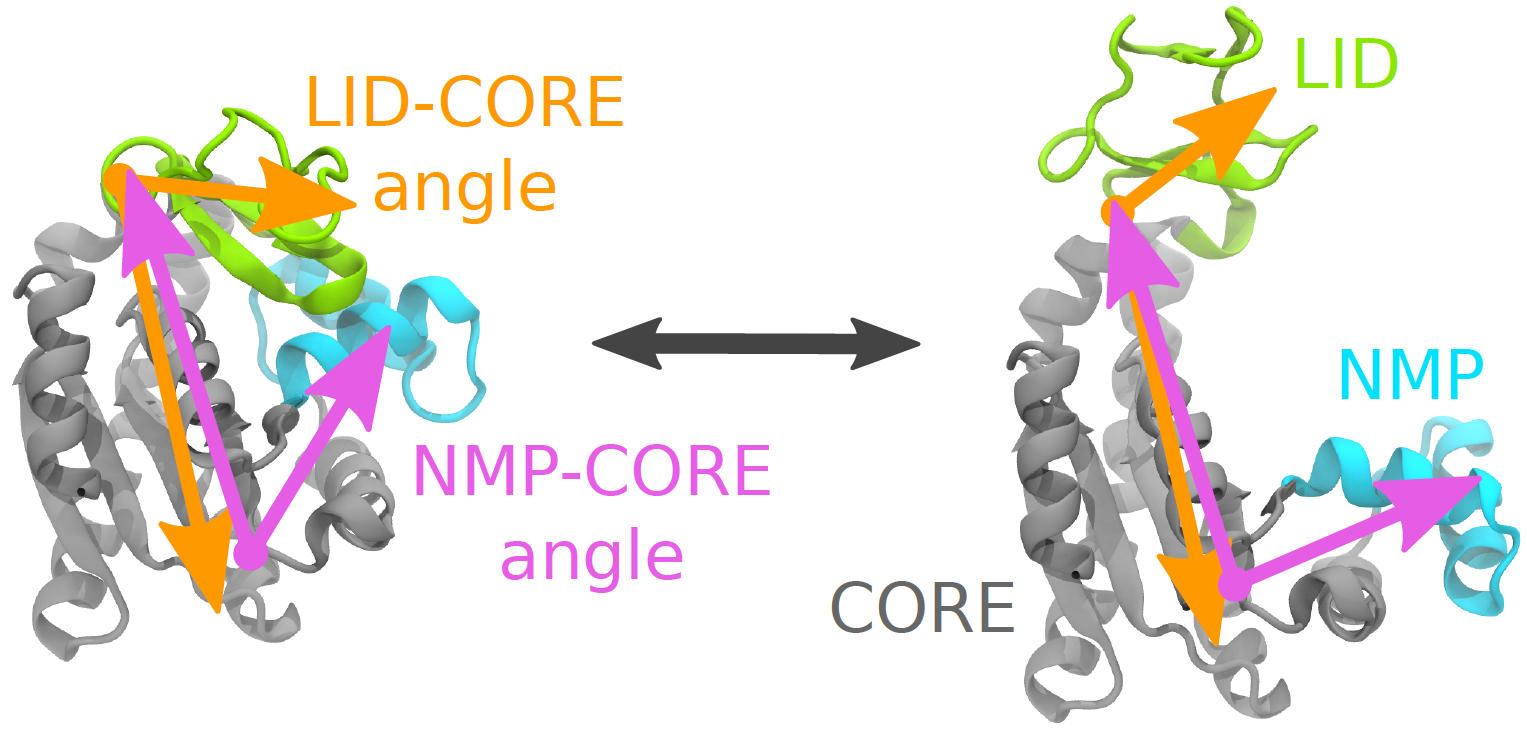
Collective variables: angle coordinates describing the AdK conformational transition. The enzyme AdK undergoes a large conformational change between a closed (left) and open (right) conformation, during which the LID domain (yellow) and NMP domain (blue) move relative to the CORE (gray). One choice of variables for quantifying the two bending motions is the LID-CORE angle (orange) and the NMP-CORE angle (purple).
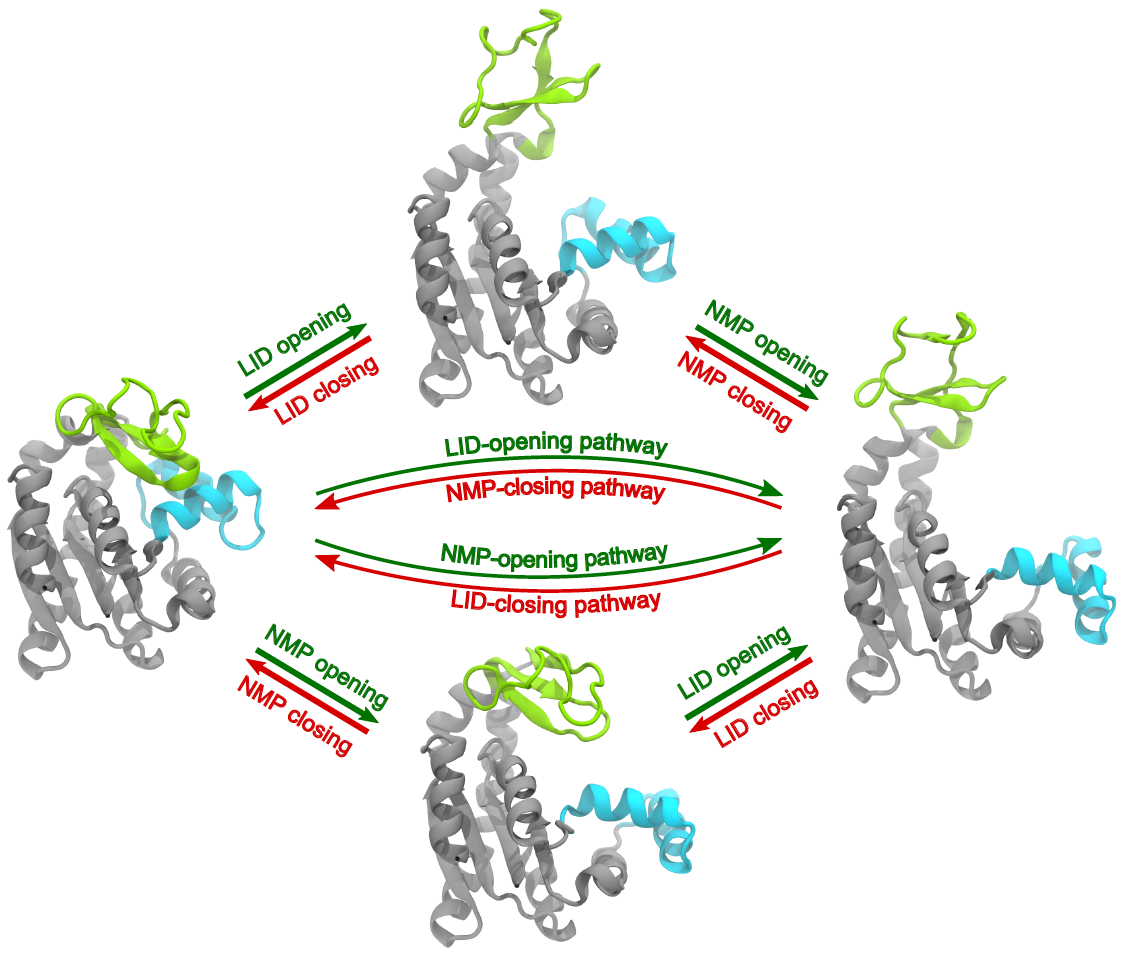
AdK conformational transition pathways. Two plausible pathways for the conformational change between a closed (left - represented by PDB ID 1ake) and open (right - represented by PDB ID 4ake) conformation, during which the LID domain (green) and NMP domain (blue) move sequentially. The top path in the closed to open direction (green arrows) consists of the LID-opening first, placing AdK in one possible intermediate state (top - represented by PDB ID 2ak3), followed by opening of the NMP domain to the open state. The bottom path in the closed to open transition is defined by NMP-opening preceding the LID-opening, with a LID-closed/NMP-open intermediate structure (bottom - represented by PDB ID 1ak2). The red arrows show the two domain closing orders for the open to closed transition.
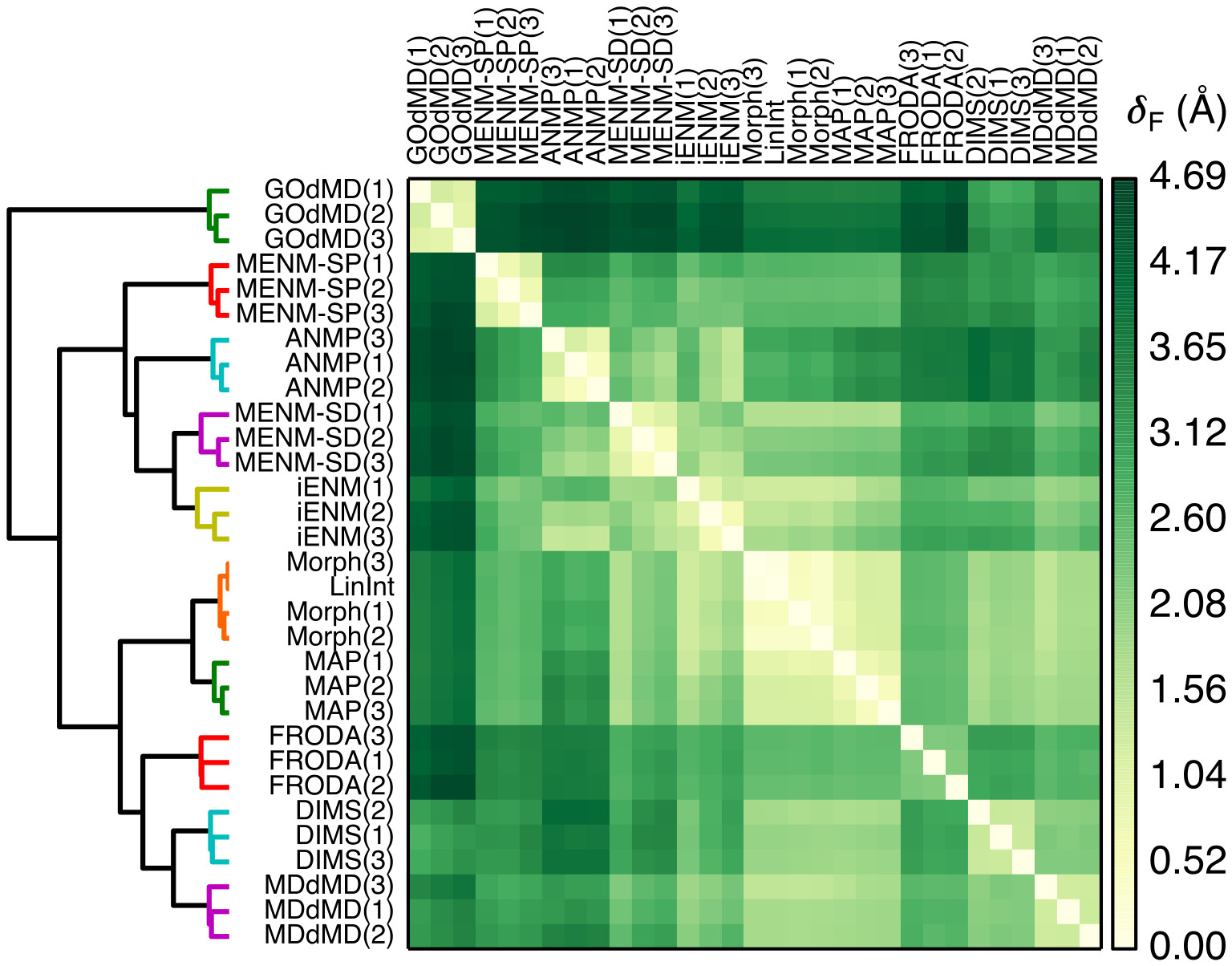
We introduced a novel method in this review for quantifying transition path similarity using the Hausdorff and Fréchet path metrics, without relying on a priori choices of collective variables, and apply it to the comparison of AdK closed —> open transitions from various path sampling methods. (See our paper on Path Similarity Analysis: a Method for Quantifying Macromolecular Pathways.2) A powerful approach for exploratory analysis of similarity measurements between transition paths is to use clustering. We use as a visual representation a heat map-dendrogram plot of the path similarity analysis of these transition paths. Hierarchical clustering with the Ward criterion was used to cluster the Fréchet distance matrix generated by pairwise Fréchet distance measurements between the paths. Distances are in Å rmsd (between structures in the paths); large deviations between paths are dark green, while small deviations are lighter.
A heat map-dendrogram representation of path similarity analysis of AdK closed to open transition paths generated by different methods. Hierarchical clustering with the Ward criterion was used to cluster the Fréchet distance matrix generated by pairwise Fréchet distance measurements between the paths. Distances are in Å rmsd (between structures in the paths); large deviations between paths are dark green, while small deviations are lighter.
References
- a Seyler, S. L. and Beckstein, O. Sampling large conformational transitions: adenylate kinase as a testing ground. Mol. Simul. 40 (10-11), 855-877 (2014). doi: 10.1080/08927022.2014.919497 . Also available from the ASU Digital Repository under accession number 27127
- a Seyler SL, Kumar A, Thorpe MF, Beckstein O (2015) Path Similarity Analysis: A Method for Quantifying Macromolecular Pathways. PLoS Comput Biol 11 (10): e1004568. doi: 10.1371/journal.pcbi.1004568



Discuss: “Sampling macromolecular transitions”