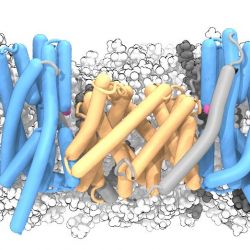
Molecular basis of ion translocation in sodium/proton antiporters
The transport of ions across the cell membrane is an essential process in all cells. However, although we know the proteins responsible for this transport, for many it is still unclear how they perform this role. One such class of proteins are the sodium/proton antiporters: transporters that exchange protons for sodium ions. These transporters are important drug targets because their dysfunction is linked to a range of human diseases including cancer, cardiovascular pathophysiologies, and autism.
In an effort to understand how these proteins transport ions, we focused on a bacterial transporter named NapA. This protein is similar to the human proteins, but it’s one of the few for which we have experimental structures. In our previous work we hypothesized that NapA could change shape dramatically as part of the transport process1.
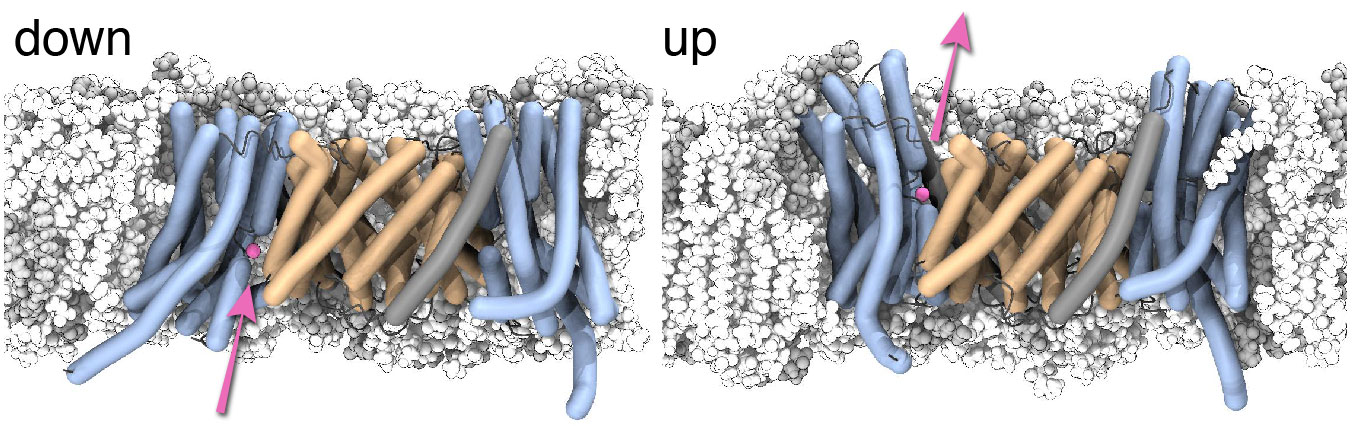
In our new paper in NSMB2, we now have conclusive evidence that NapA transports ions by a large movement of one part of the protein. The movement resembles the movement of an elevator, where the ion enters the protein on one side, then the elevator portion of the protein moves, and the ion exits on the other side. In the image below the ion is magenta, the protein is shown in cartoon representation in wheat and blue, the lipids of the membrane are white/gray. The “core” domain (blue) moves from the down position to the up position while the “dimer” domain (wheat) anchors the transporter to the membrane. The ion binds inside the core domain and is shuttled through the membrane.
Two images from MD simulations of NapA in the inward facing (left, down) and outward facing (right, up) conformation. A sodium ion (magenta) can bind to the inward facing conformation. The "core" domain (blue) can move up in an elevator-like conformational transition and the ion can exit on the other side.
(The work on NapA comes from an ongoing collaboration between the experimental labs of David Drew at the University of Stockholm (Sweden), Alexander Cameron at the University of Warwick (UK) and our lab.)
Elevator-type movements are wide-spread in alternating access transporters
In the same issue of NSMB, the groups of Joe Mindell and Lucy Forrest report on a combined experimental/modelling study of the secondary dicarboxylate transporter VcINDY 3. VcINDY is not related to NapA but it also forms a dimer and the new study shows that it also undergoes a large elevator-like motion. It thus joins a growing number of confirmed elevator-transporters such as the aspartate transporter GltPh4 (which was the first transporter for which the phrase “elevator movement” was coined), homologs of ASBT5 and the citrate transporter CitS6.
The growing realization that large vertical movements of fairly rigid domains (often combined with rotations of the moving domain) are more wide-spread in alternating-access transporters than previously thought was recently summarized in a NSMB News & Views article by Ryan and Vandenberg7.
References
- a C. Lee, H.J. Kang, C. von Ballmoos, S. Newstead, P. Uzdavinys, D. L. Dotson, S. Iwata, O. Beckstein, A.D. Cameron, and D. Drew. A two-domain elevator mechanism for sodium/proton antiport. Nature 501 (2013), 573–577. doi: 10.1038/nature12484
- a Coincon, P. Uzdavinys, E. Nji, D. L. Dotson, I. Winkelmann, S. Abdul-Hussein, A. D. Cameron, O. Beckstein, and D. Drew. Crystal structures reveal the molecular basis of ion-translocation in sodium/proton antiporters. Nature Struct. Mol. Biol. 23 (2016), 248–255. doi: 10.1038/nsmb.3164
- a Mulligan, C. Fenollar-Ferrer, G. A. Fitzgerald, A. Vergara-Jaque, D. Kaufmann, Y. Li, L. R. Forrest, and J. A. Mindell. The bacterial dicarboxylate transporter VcINDY uses a two-domain elevator-type mechanism. Nat Struct Mol Biol, 23 (2016), 256–262, doi: 10.1038/nsmb.3166
- a N. Reyes, C. Ginter, and O. Boudker. Transport mechanism of a bacterial homologue of glutamate transporters. Nature, 462 (2009), 880–885. doi: 10.1038/nature08616
- a Zhou, E. J. Levin, Y. Pan, J. G. McCoy, R. Sharma, B. Kloss, R. Bruni, M. Quick, and M. Zhou. Structural basis of the alternating-access mechanism in a bile acid transporter. Nature, 505 (2014), 569–573. doi: 10.1038/nature12811
- a D. Wöhlert, M.J. Grötzinger, W. Kühlbrandt, and Ö. Yildiz. Mechanism of Na+-dependent citrate transport from the structure of an asymmetrical CitS dimer. eLife, 4 (2015), e09375. doi: 10.7554/eLife.09375
- a M. Ryan and R. J. Vandenberg. Elevating the alternating-access model. Nat Struct Mol Biol, 23 (2016), 187–189. doi: 10.1038/nsmb.3179


Discuss: “Molecular basis of ion translocation in sodium/proton antiporters”