Cryo-electron microscopy and Simulations
Cryo-electron microscopy has become a powerful new structural technique to obtain atomic resolution structures of membrane proteins. By combining the experimental structures with molecular dynamics simulations of the protein in a realistic environment including a membrane and solvent (water and ions), new insights regarding the protein function can be obtained that are not available from a single technique alone.
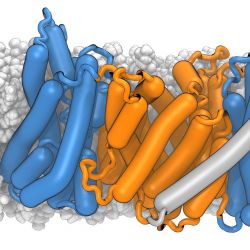
The structure of a membrane protein, the mammalian sodium/proton antiporter NHE9, was solved by cryo-electron microcsopy by our collaborators. In order to better understand its interaction with the membrane we simulated the NHE9 dimer in a POPC:cholesterol membrane. The dimerization domain is shown in orange while the core domains, where a sodium ion can bind, are depicted in blue. The story behind this image is described under Structure and function of the mammalian sodium/proton exchanger NHE9.
Ricky Sexton created the image (which made the cover of EMBO J) in VMD and rendered with the Tachyon Raytracer with ambient occlusion lighting. The protein “sausage” representation was made with the Bendix VMD plugin.


