
Google Summer of Code 2023: MDAnalysis
MDAnalysis is looking for undergraduate/graduate students or anyone new to open source to participate in Google Summer of Code (GSoC) 2023. Application window closes April 4, 2023 – 18:00 UTC.

MDAnalysis is looking for undergraduate/graduate students or anyone new to open source to participate in Google Summer of Code (GSoC) 2023. Application window closes April 4, 2023 – 18:00 UTC.

We have a Postdoctoral Scholar position available to start as soon as possible (summer 2022) to work on the MDAnalysis library to launch the new MDAKits eco-system of analysis packages. The position can be performed remotely. The initial deadline to apply is July 10, 2022.

MDAnalysis is looking for students or anyone new to open source to participate in Google Summer of Code (GSoC) 2022. Application window closes April 19, 2022 at 18:00 (UTC).

The Chan Zuckerberg Initiative recognized our MDAnalysis software with an Essential Open Source Software for Science (EOSS) grant. ASU News published an interview with Oliver about what the grant means for the MDAnalysis project.

Alia Lescoulie is a chemistry undergraduate at Cal Poly in San Luis Obispo. Her research has consisted of applying computational methods to different problems in chemistry. She participated in the 2021 SPIDAL REU and worked on developing tools for analyzing molecular dynamics data.

MDAnalysis is looking for students to participate in Google Summer of Code (GSoC) 2021. Application window closes April 13, 2021 at 18:00 (UTC).

The Beckstein Lab offers fully funded ten-week virtual research programs in computational biophysics for highly motivated undergraduate students. This is a NSF-sponsored Research Experience for Undergraduates (REU). Deadline for applications is March, 7 2021.
Due to the ongoing COVID-19 pandemic, the REU will be performed remotely and will not include travel to ASU.*

Edis Jakupovic was in the Physics Ph.D. program at Arizona State University. In 2021, he graduated with a bachelors degree in Physics and Biochemistry from ASU. In spring 2025, Edis decided to pursue other opportunities beyond the physics PhD.

MDAnalysis is looking for students to participate in Google Summer of Code (GSoC) 2020. Application window closes March 31, 2020 at 18:00 (UTC).

The Beckstein Lab offers a fully funded ten-week virtual research program in computational biophysics for a highly motivated undergraduate student. This is a NSF-sponsored Research Experience for Undergraduates (REU). Deadline for applications is April, 30 2020.
Due to the ongoing COVID-19 pandemic, the REU will be performed remotely and will not include travel to ASU.*

Nikolaus Awtrey is a physics graduate at Arizona State University and a visiting researcher at Beckstein Lab.

MDAnalysis is looking for technical writers to participate in the inaugural Google Season of Docs (GSoD) 2019. Application window closes June 28, 2019 at 18:00 UTC.

The Beckstein Lab offers a fully funded ten-week research program in computational biophysics for a highly motivated undergraduate student. This is a NSF-sponsored Research Experience for Undergraduates (REU). Deadline for applications is April, 14 2019.

MDAnalysis is looking for students to participate in Google Summer of Code (GSoC) 2019. Application window closes April 9, 2019.

Henry Mull is an undergraduate chemistry student at Cal Poly San Luis Obispo. He participated in the Summer 2018 REU Program with the Beckstein Lab and worked on new analysis tools for MDAnalysis.

This summer’s mini-workshop series is designed to provide introductions to various topics of interest to lab members. Weekly topics are chosen based on interest levels of lab members.

The Beckstein Lab offers a fully funded ten-week research program in computational biophysics for a highly motivated undergraduate student. This is a NSF-sponsored Research Experience for Undergraduates (REU). Deadline for applications is May 30, 2018.

MDAnalysis is looking for students to participate in Google Summer of Code (GSoC) 2018. Application window closes March 27, 2018.

The Beckstein Lab offers a fully funded ten-week research program in computational biophysics for a highly motivated undergraduate student. This is a NSF-sponsored Research Experience for Undergraduates (REU). Deadline for applications is May 5, 2017.

MDAnalysis is looking for students to participate in Google Summer of Code (GSoC) 2017. Application window is March 20 – April 3, 2017.

This year at SciPy 2016 in Austin, TX, David Dotson presented his work on datreant : Persistent, Pythonic Trees for Heterogeneous Data and Oliver Beckstein talked about MDAnalysis : A Python Package for the Rapid Analysis of Molecular Dynamics Simulations. Video recordings of both talks are available here.

Members of the Becksteinlab will present talks at SciPy 2016. This year the biggest conference on Python in science takes place from July 11-17 in Austin, TX. We will talk about datreant and MDAnalysis, get inspired, meet old and new friends, and hand out MDAnalysis stickers.

Robert Delgado is an undergraduate pursuing a Bachelor’s of Science in Cornell’s Applied and Engineering Physics Program who spent the summer of 2016 in the Beckstein lab as a REU student.

The Beckstein Lab offers a fully funded ten-week research program in computational biophysics for a highly motivated undergraduate student. This is a NSF-sponsored Research Experience for Undergraduates. Deadline for applications is May 30, 2016.

MDAnalysis is looking for students to participate in Google Summer of Code (GSoC) 2016. Start with the blog post MDAnalysis: Google Summer of Code 2016 .
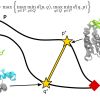
Transition pathways in high dimensional spaces, such as the ones produced by advanced algorithms to sample large conformational changes in macromolecules, are difficult to analyze quantitatively. We introduce a method named Path Similarity Analysis (PSA) that enables us to quantify the similarity between two arbitrary paths and extract the atomic-scale determinants responsible for their differences. PSA is implemented in the MDAnalysis library.

The CECAM Macromolecular simulation software workshop will be held from Mon 12 Oct, 2015 to Sat 24 Oct 2015 at Forschungszentrum Jülich, Germany. David Dotson and Oliver Beckstein will participate and will teach how to use MDAnalysis productively.

The Beckstein Lab offers a fully funded ten-week research program in computational biophysics for a highly motivated undergraduate student. This is a NSF-sponsored Research Experience for Undergraduates. Deadline for applications is May 31, 2015.

The Lab is a partner on the large NSF-sponsored SPIDAL project (“Middleware and High Performance Analytics Libraries for Scalable Data Science”) that was announced as part of a larger program for Laying the groundwork for data-driven science. The lab joins with partners at Indiana University, Emory University, Rutgers University, University of Kansas, University of Utah and Virginia Tech.

MDAnalysis is an open source, versatile, object-oriented Python library for analyzing molecular dynamics trajectories. It makes it easy to access trajectory data from Python code by interfacing trajectory readers (and writers) with NumPy arrays and to select atoms via a expressive selection syntax. The CHARMM/NAMD, Amber, Gromacs trajectory formats are all supported as well as PDB, GRO, CRD, PQR, and a range of others.